The influence of environmental change on genetic diversity across spatial and
taxonomic scales
Connor French, PhD Candidate
Pre-defense seminar, 2024-06-04
The Graduate Center
City University of New York




The Earth is constantly changing
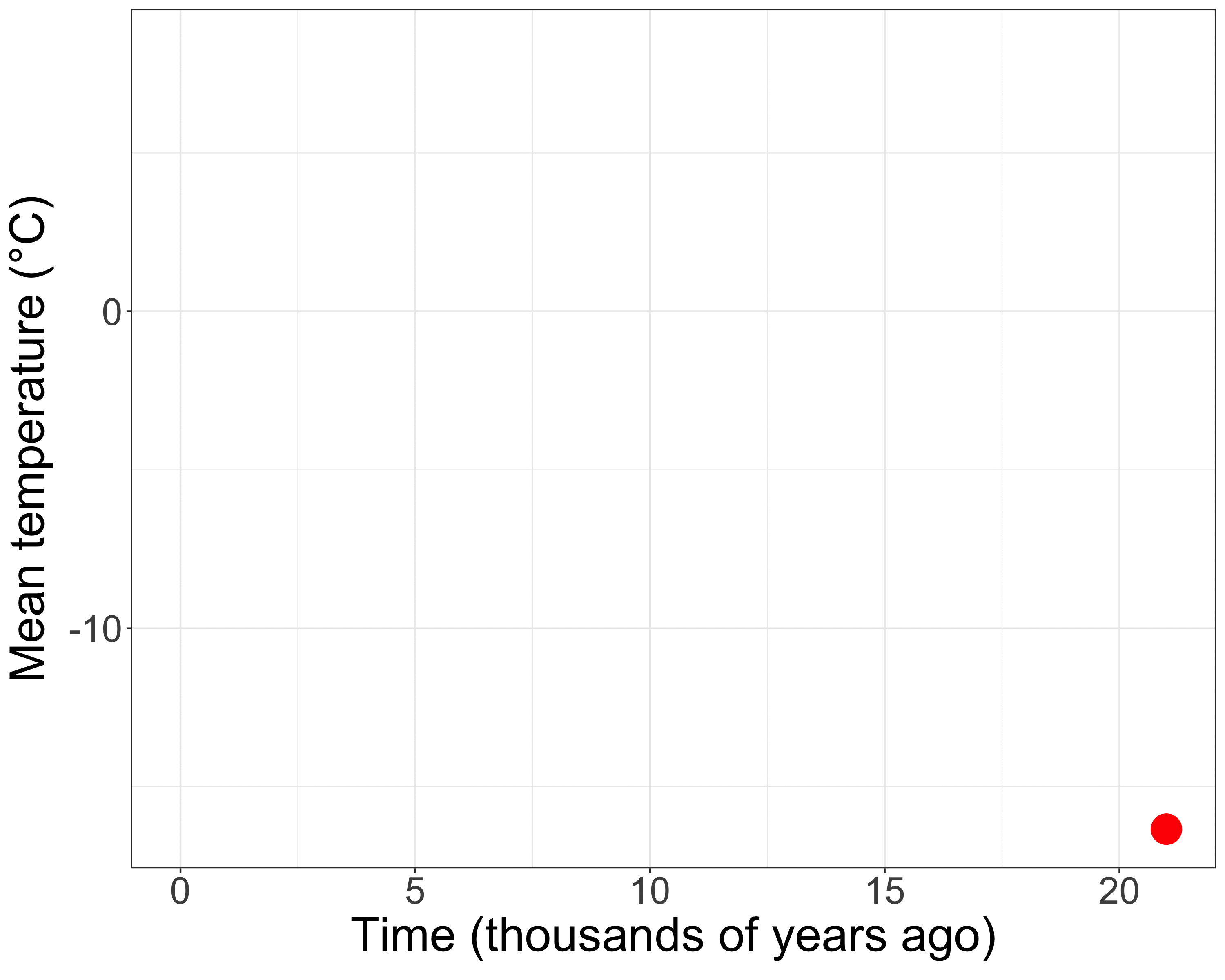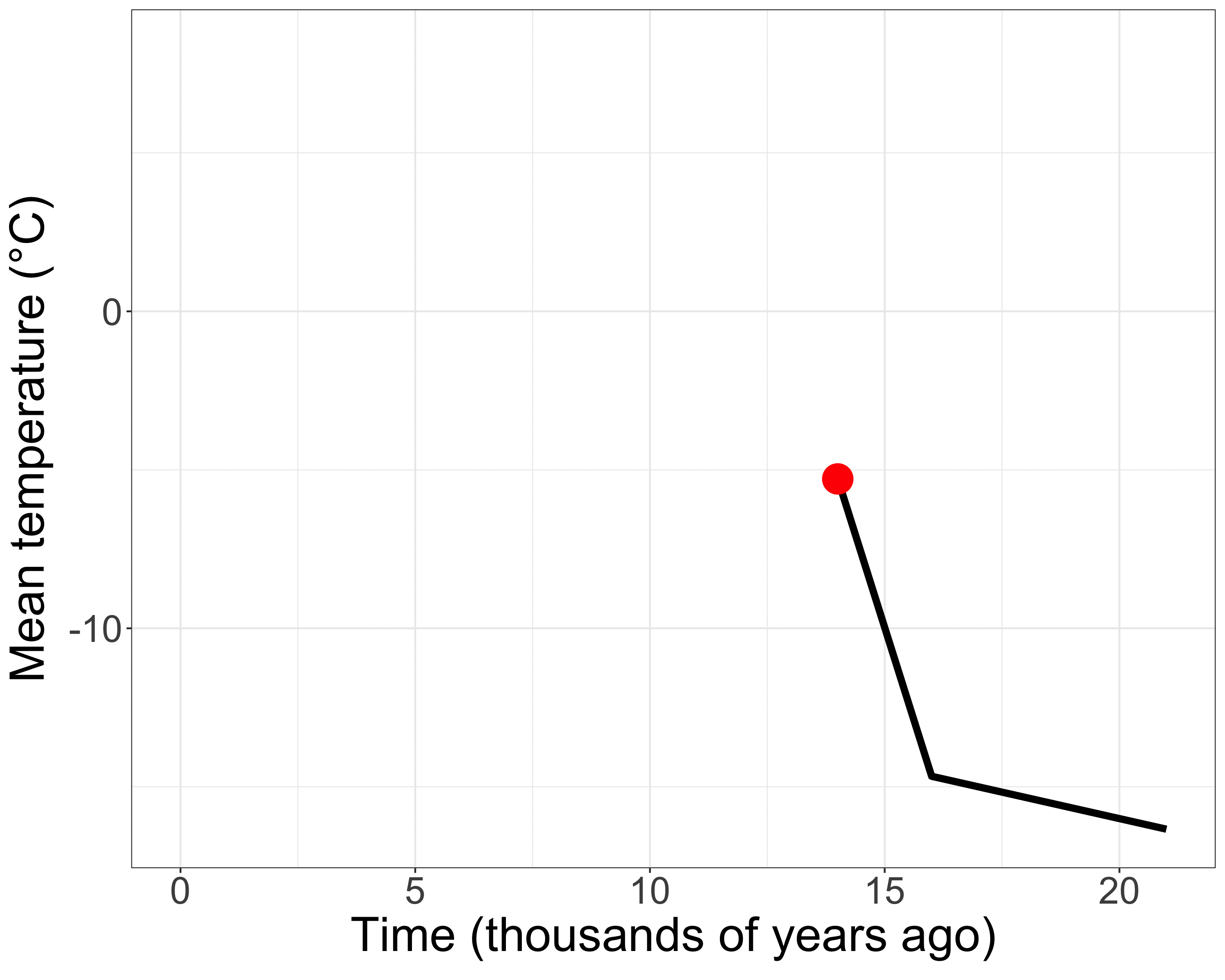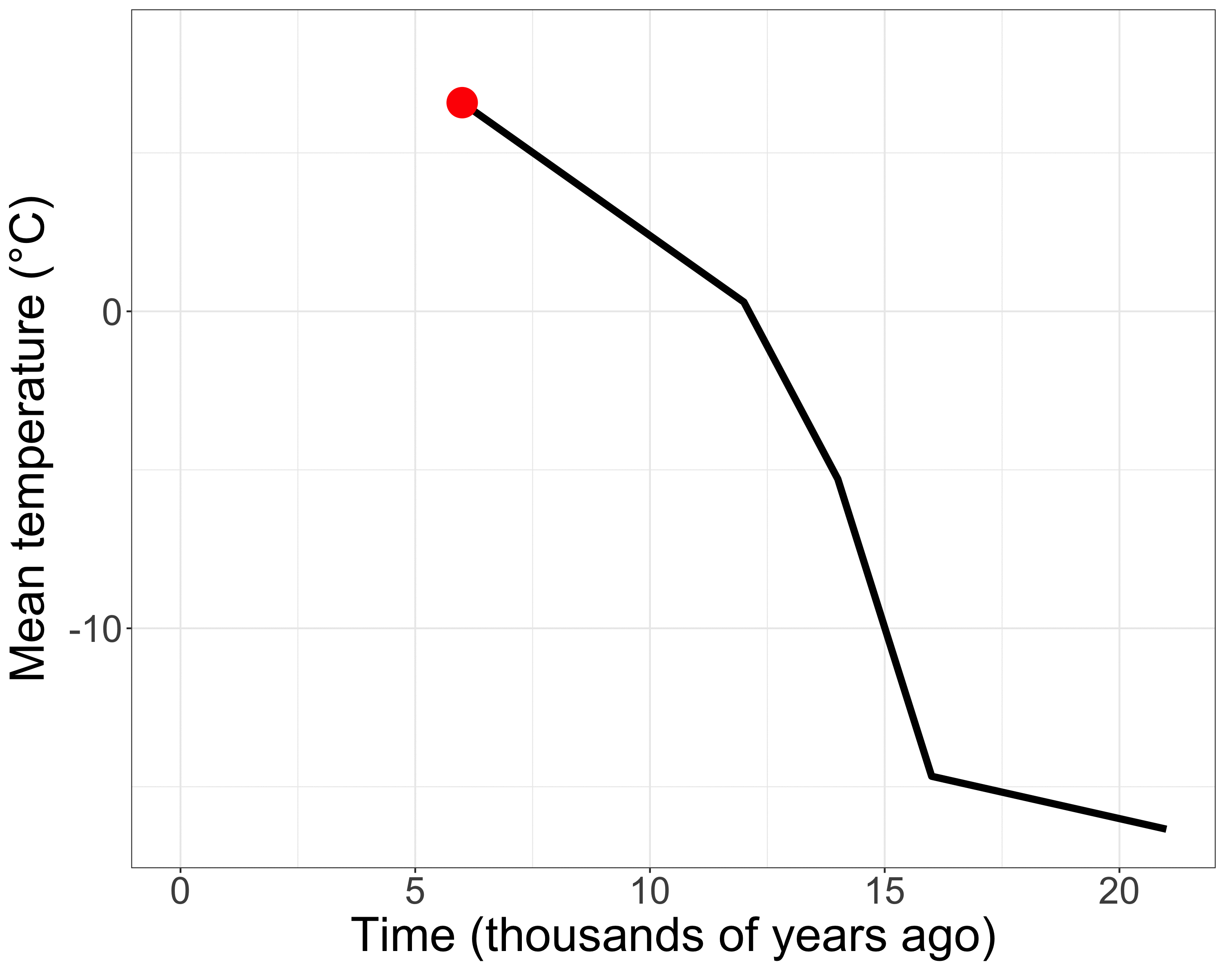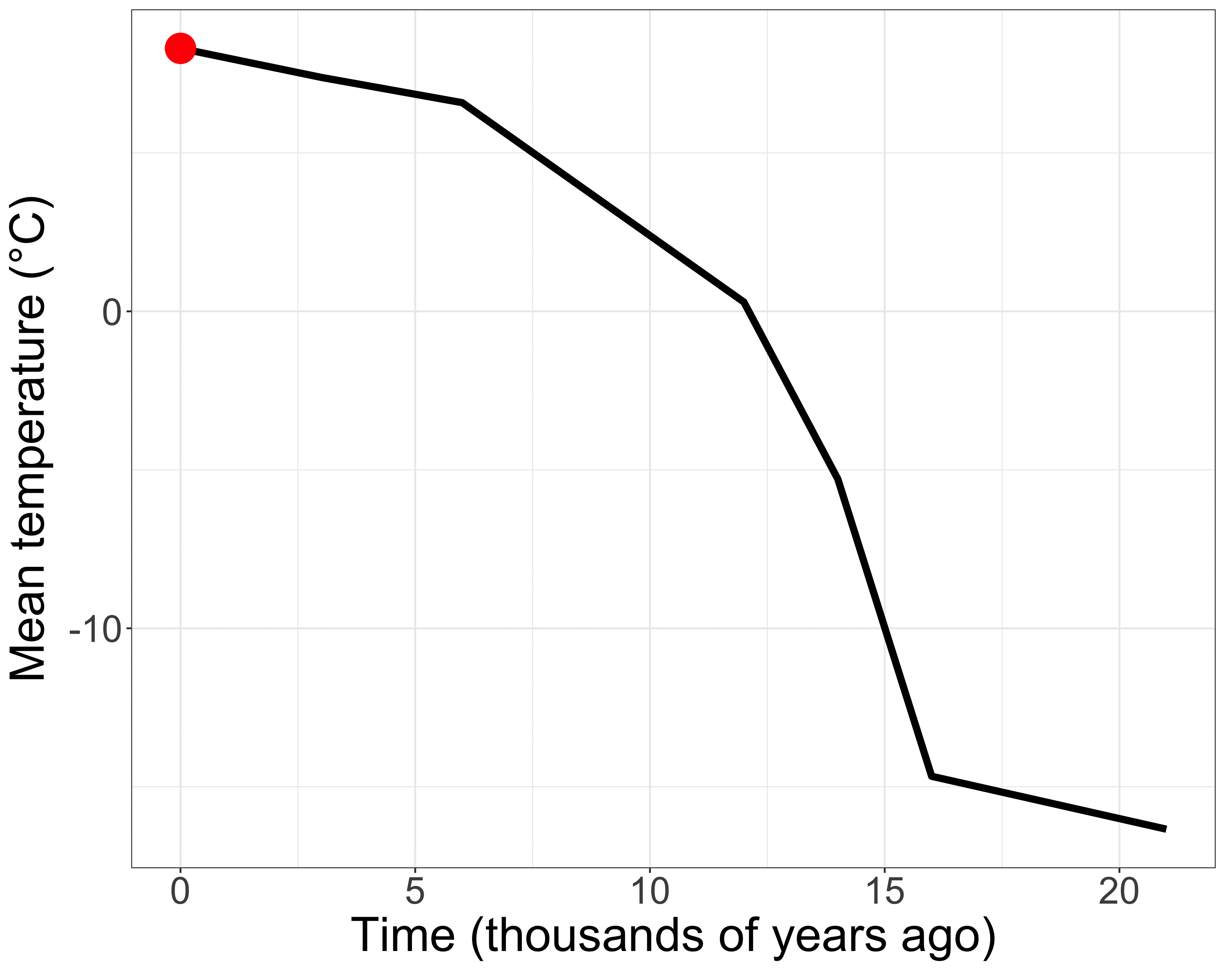
Populations shrink or expand…
Fragment and reconnect…
Adapt or go extinct
Genetic diversity contains signatures of that change
Video from: https://bedford.io/projects/coaltrace/
Genetic diversity can tell stories about
Species
and Communities
1
Understanding these stories is especially urgent
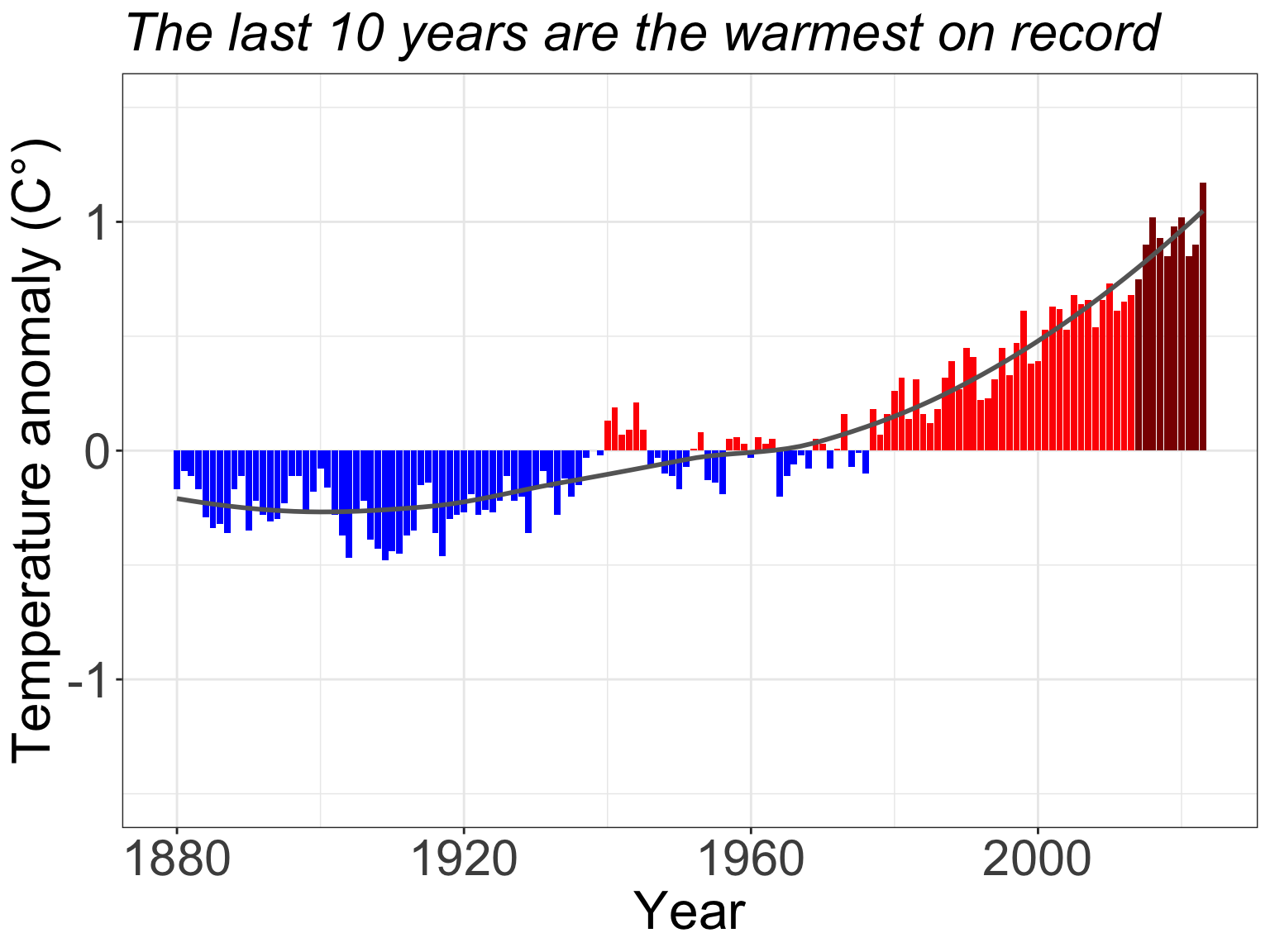
Understanding these stories is especially urgent
Ectotherms are linked to their environments


I investigate global and regional patterns of genetic diversity in two groups of ectotherms, insects and lizards, to understand the relationship between environmental change and genetic diversity, from populations to assemblages.
My questions
1. What is the relationship between genetic diversity and environmental change in insects?
2. What can genetic diversity convey about the processes underlying species responses to environmental change?
3. How can I make integrative modeling of species responses to environmental change more accessible?
Global determinants of insect mitochondrial genetic diversity

A global perspective is necessary
Ceballos and Ehrlich 2006 PNAS 10.1073/pnas.0609334103
Insects comprise over 93% of the planet’s described animal diversity
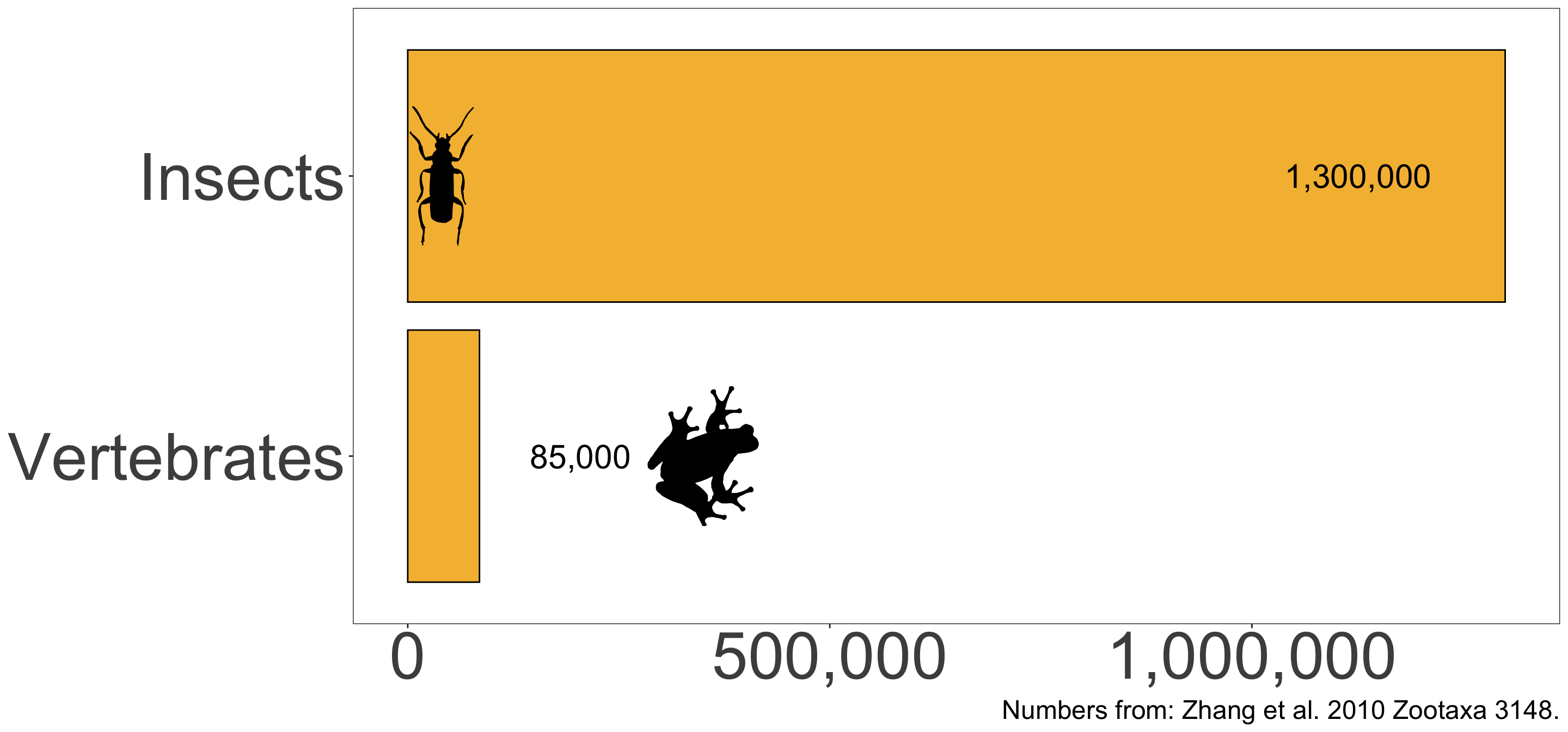
Global biodiversity patterns remain undocumented for most insects
- Scientists have relied on species richness or species diversity
- Describing and cataloging the world’s insect diversity is a monumental task
- What do we do?
Genetic diversity is a promising biodiversity metric
- It does not rely on species identity for calculation
- It conveys historical information
- macrogenetics is an emerging field
Leigh et al. 2022. Nat. Rev. Genet. 10.1038/s41576-021-00394-0
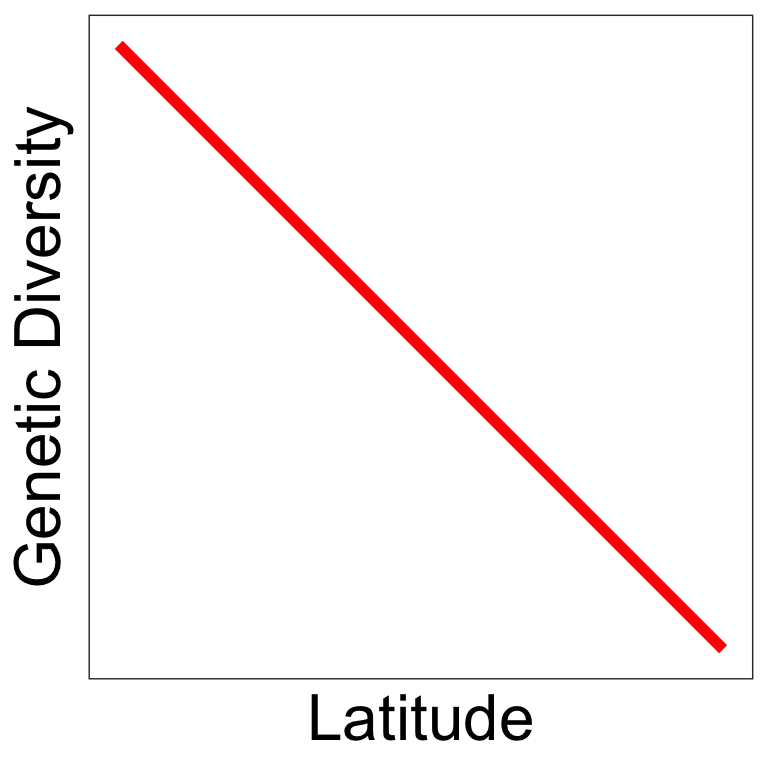
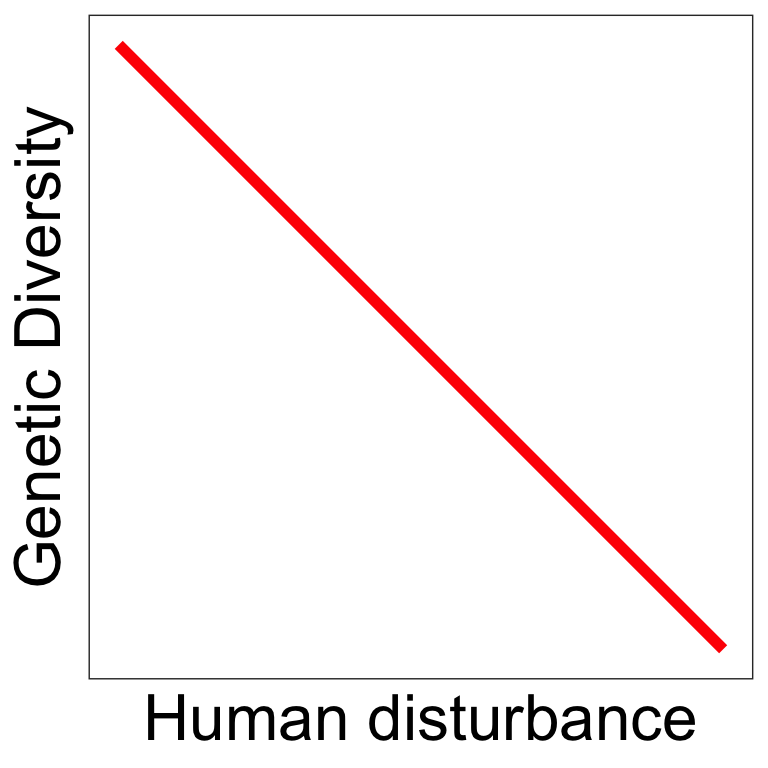
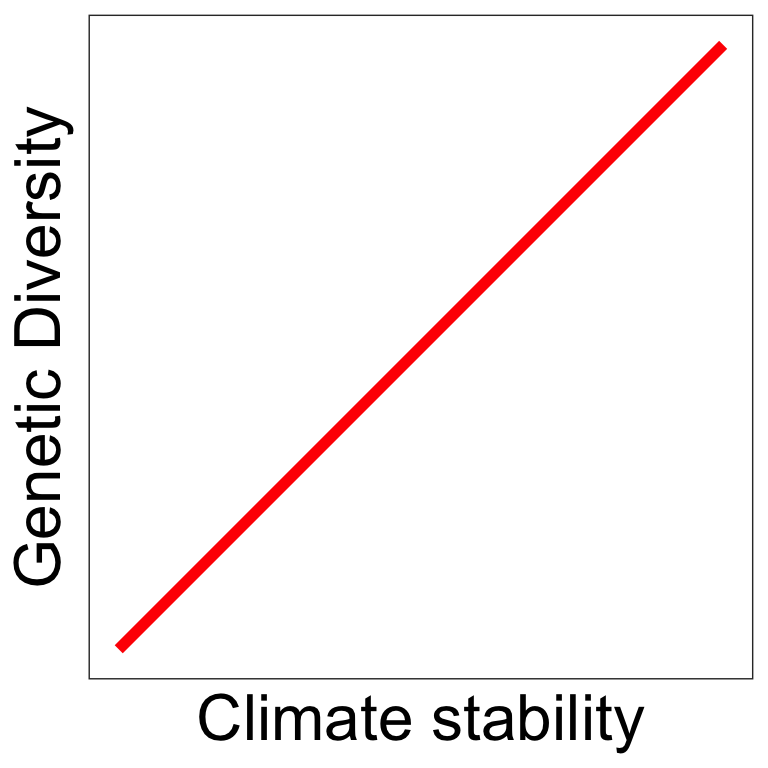
Some predictions follow those from vertebrates
Latitudinal diversity gradient 1
Human disturbance 2
Climate stability 3
I compiled the largest animal macrogenetic dataset to date
- 2,415,415 mtDNA sequences 1
- 98,417 operational taxonomic units (OTUs) 2
I compiled the largest animal macrogenetic dataset to date
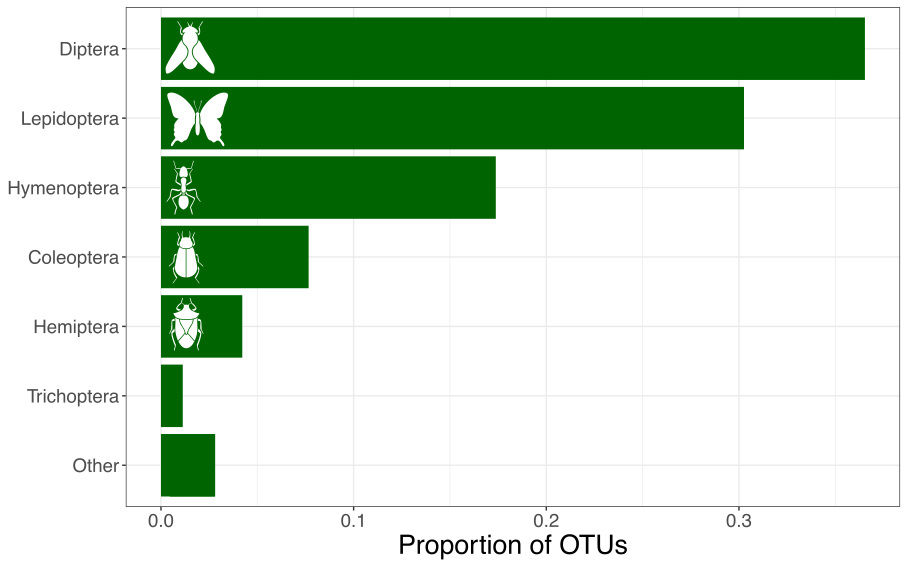
Summarizing genetic diversity
For each OTU, I calculated the mean number of pairwise differences between sequences
Genetic diversity mean (GDM) = the average genetic diversity among all OTUs
Genetic diversity evenness (GDE) = the evenness of genetic diversity across OTUs
High GDE -> more OTUs with a similar level of genetic diversity
Low GDE -> OTUs have very different levels of genetic diversity
Responses
- GDE
- GDM
Predictors
- Latitude
- Climate
- Climate stability
- Human influence
- Habitat heterogeneity
- Topography
Global maps of insect genetic diversity evenness
GDE peaks in the subtropics
Prediction
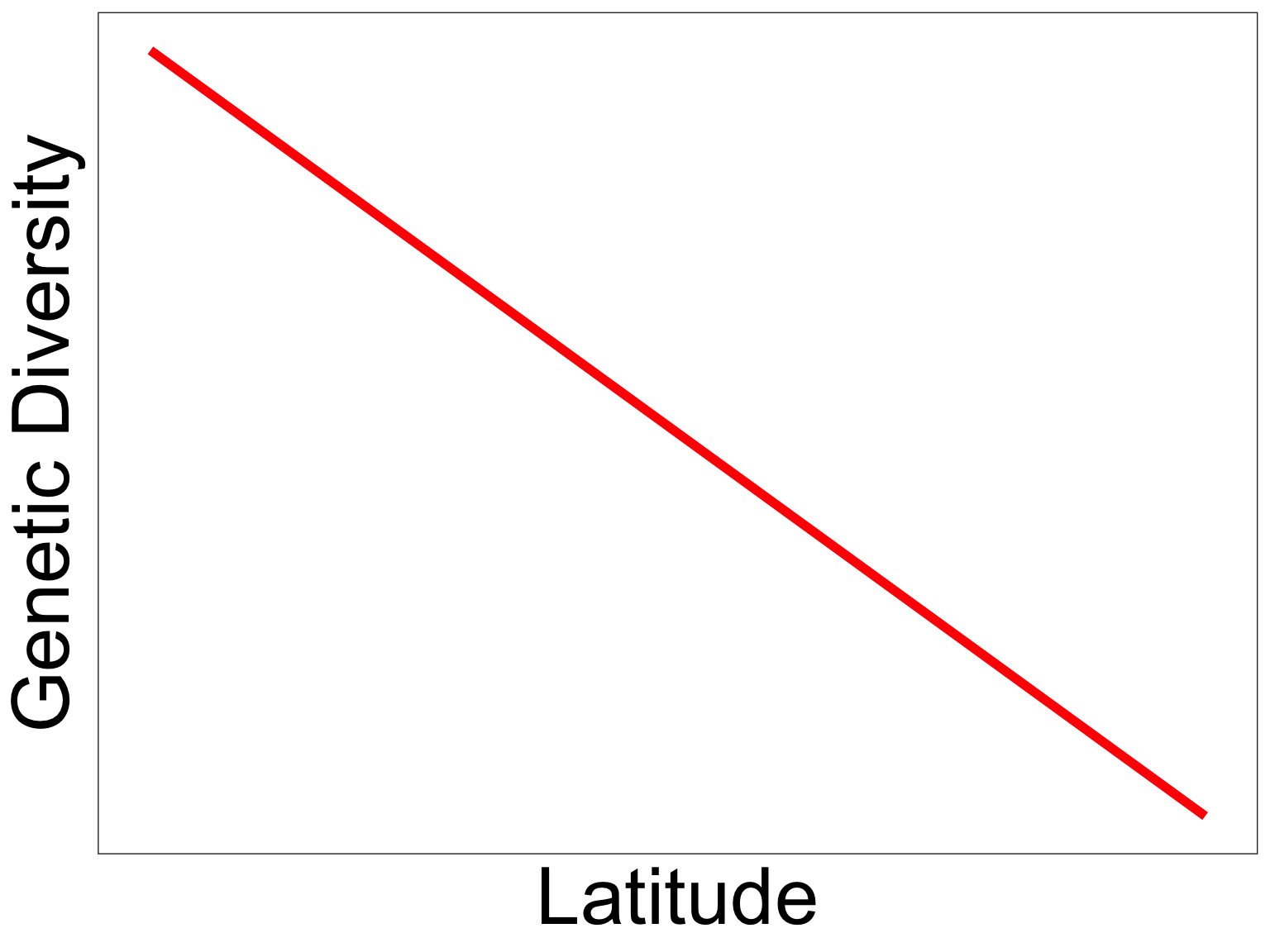
GDE peaks in the subtropics
Observation
GDE peaks in the subtropics
- Rapoport’s rule: species ranges are larger at higher latitudes 1
- Genetic diversity tends to increase with range size 2
- Pattern breaks down in previously glaciated regions
GDE is correlated with current climate and climate stability
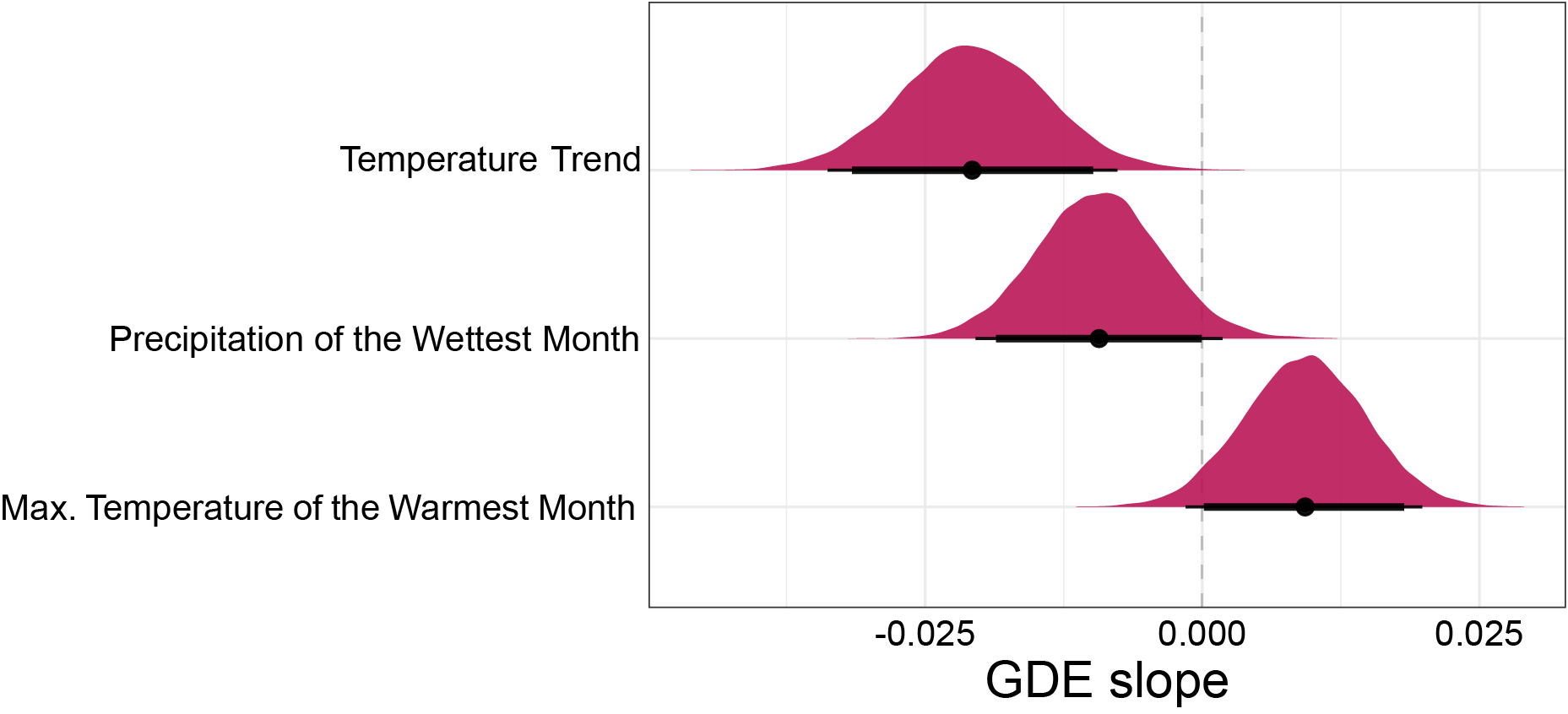
GDE is correlated with current climate and climate stability
- Climate stability hypothesis: genetic diversity increases with climate stability 1
- Evolutionary speed hypothesis: genetic diversity increases with temperature 2
- Insect diapause provides an adaptive tolerance to seasonal temperature variation 3

To sum up
- GDE peaks in the subtropics and is distinct from vertebrate patterns
- Environmental stability leads to higher levels of genetic diversity
- Seasonally warm temperatures suggest the evolutionary speed hypothesis may be at play
Demographic responses to past climate change in Brazilian Enyalius lizards using spatially-explicit coalescent modeling

Macro-scale patterns are interesting…
but what about processes?
Species are expected to respond differently to environmental change
 1
1
Species distribution models (SDMs) are useful tools
The relationship between SDM suitabilities and abundance
- While a probability may reliably predict presence, it may not linearly predict abundance
- Wider environmental tolerances may lead to higher abundances than predicted by a linear relationship in marginal habitats
Lee-Yaw et al. 2022. Ecography. 10.1111/ecog.05877
SDM suitabilities may have a non-linear relationship with abundance in high-elevation species
I investigate this relationship in two lizard species
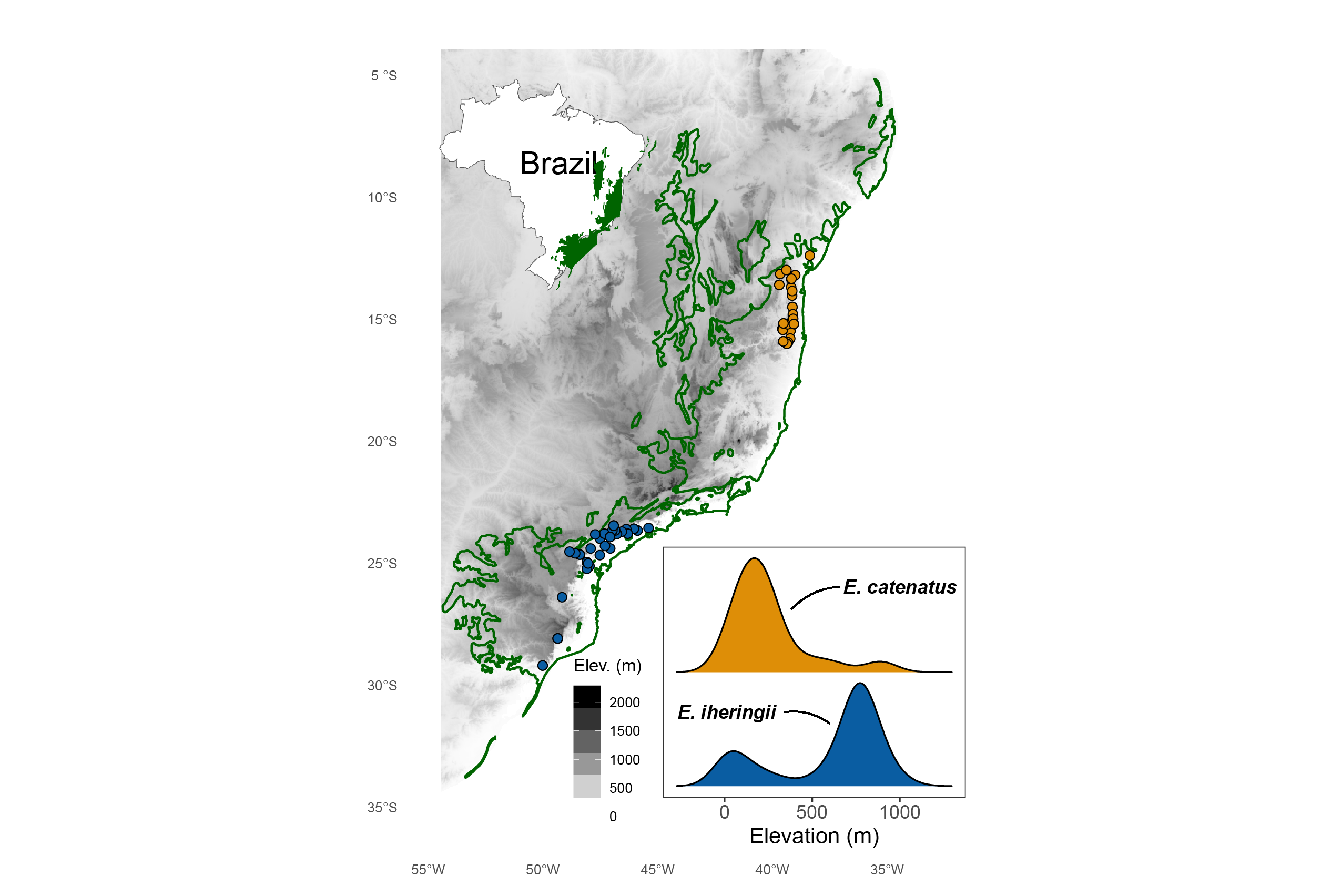

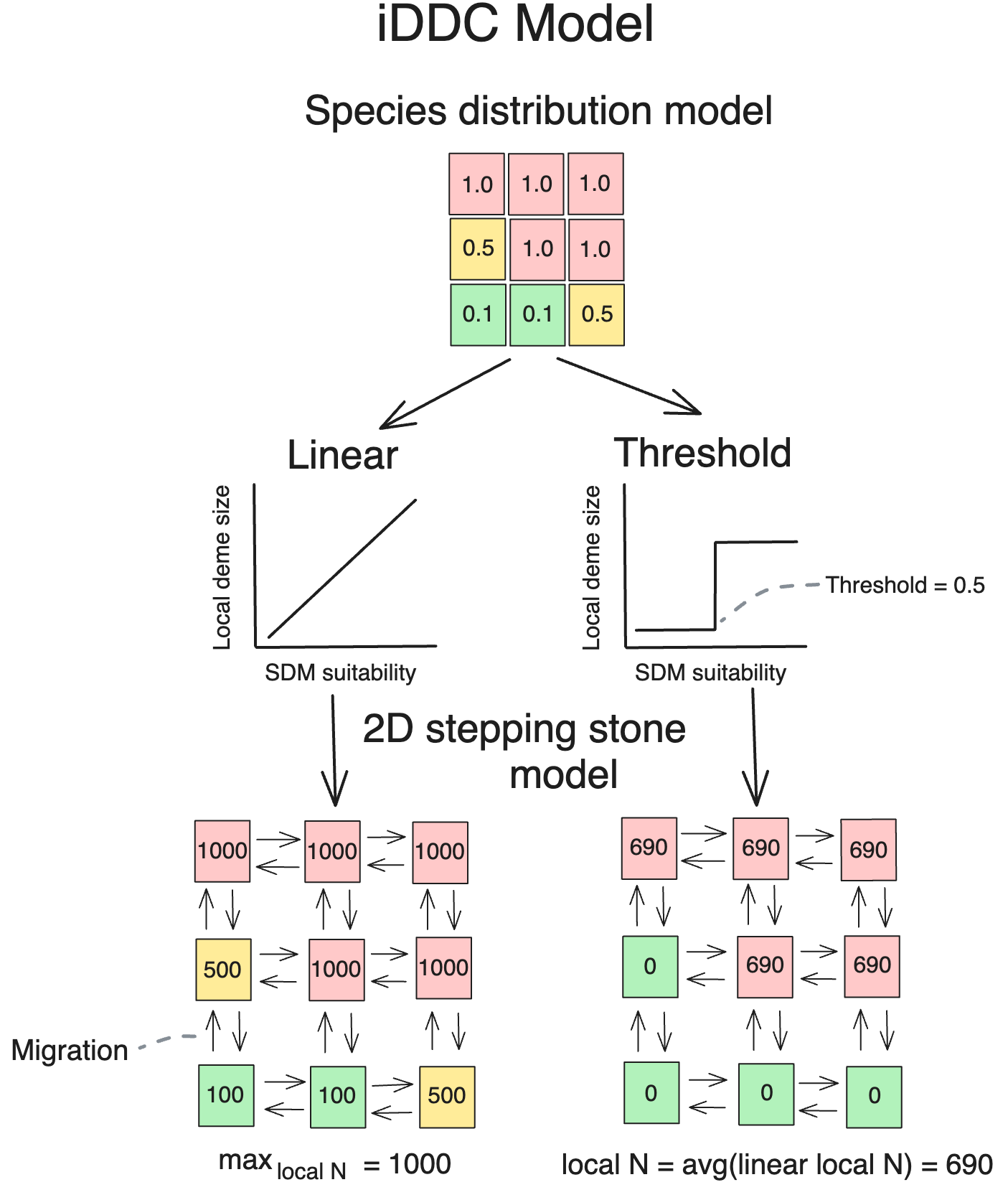
Integrative distributional, demographic, and coalescent (iDDC) modeling
iDDC models establish a link from SDM suitability to abundance to genetic diversity
Integrative distributional, demographic, and coalescent (iDDC) modeling
Distributional
- SDMs predict the distribution of suitable habitat
Demographic
- SDMs are transformed into demographic models
- local deme size and dispersal
Coalescent
- Coalescent modeling simulates the genealogy of a sample of DNA sequences backwards in time
Distributional
Demographic
Demographic/Coalescent
Predictions
Predictions
I used simulations and machine learning to test these hypotheses
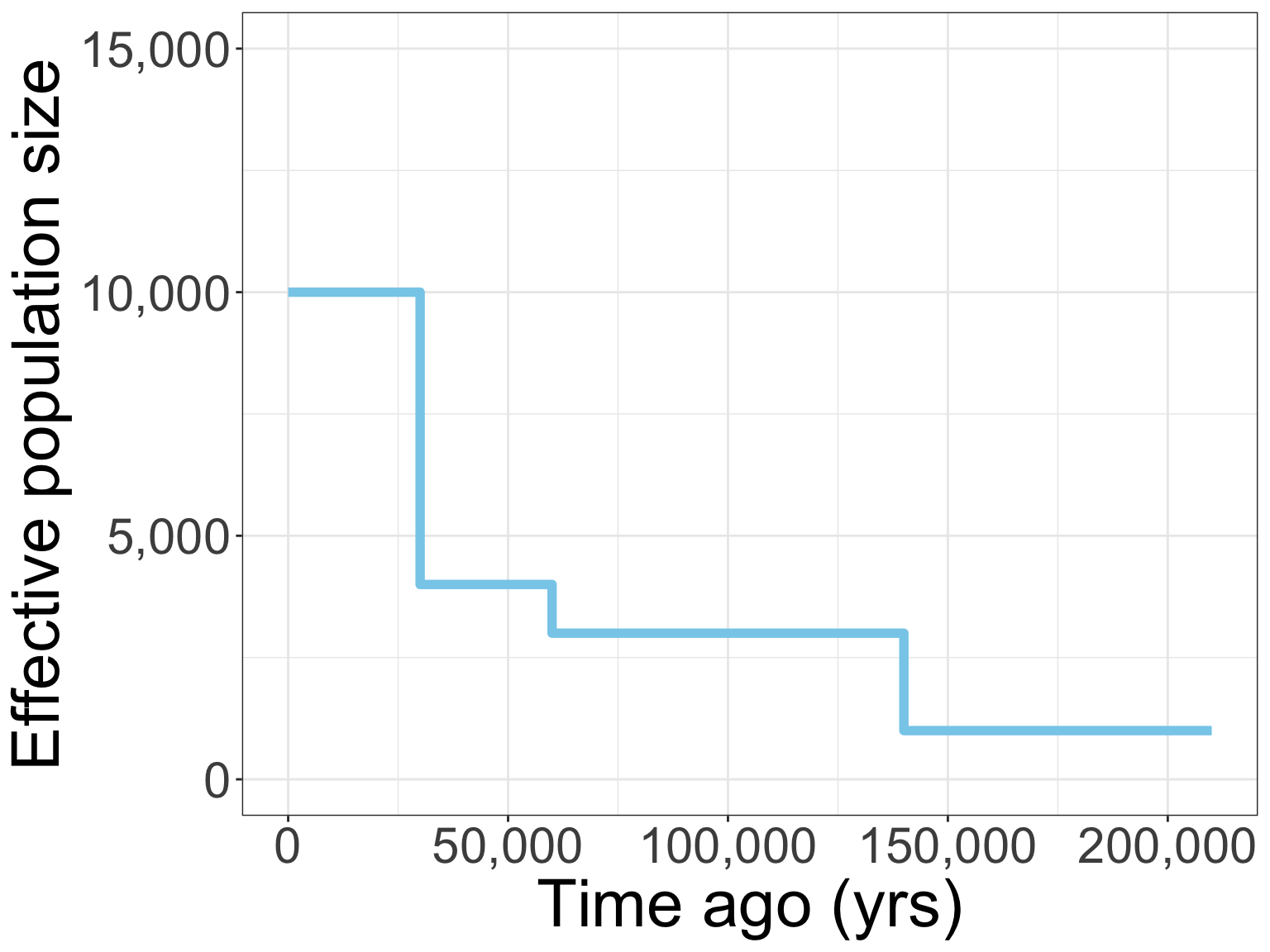
Local deme size?
Migration rate?
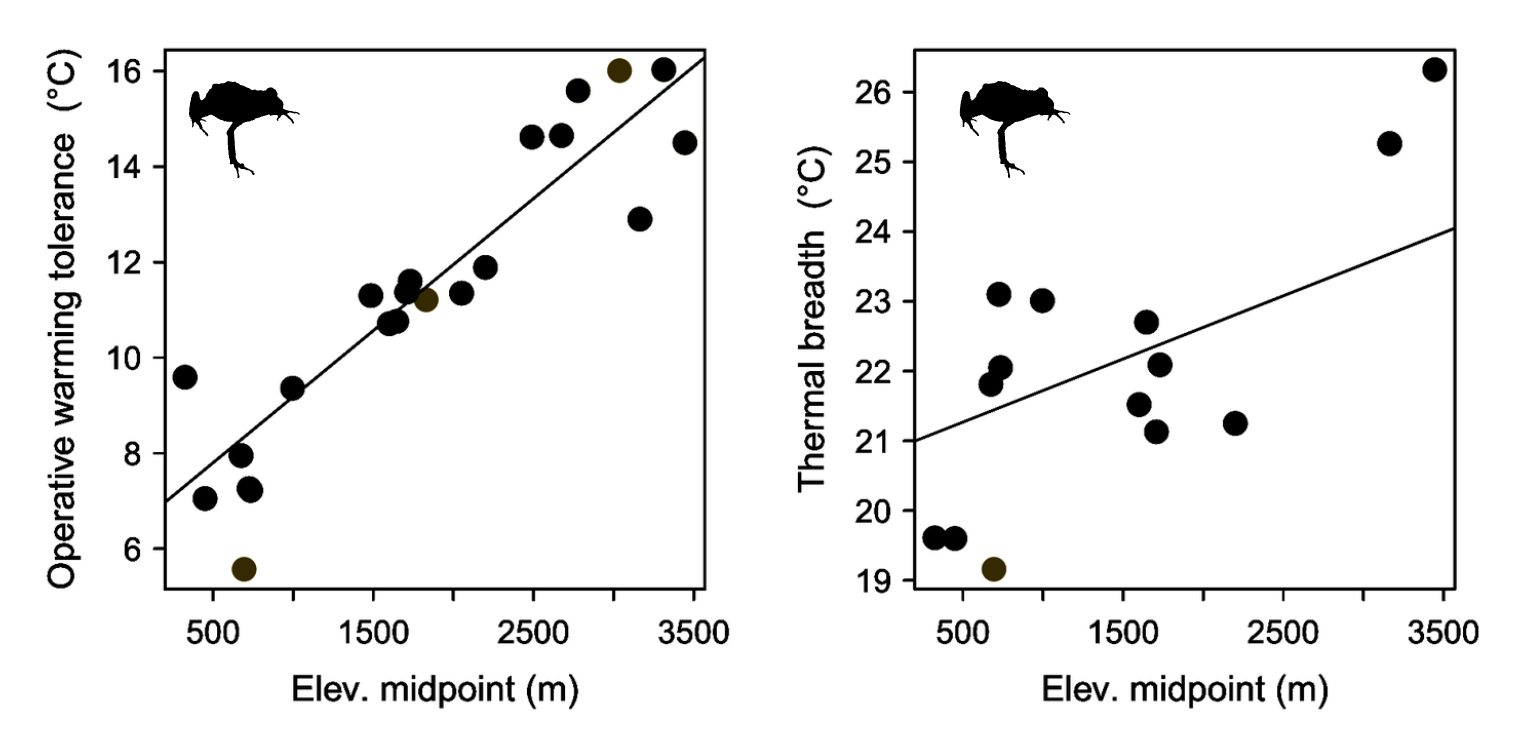
A threshold transformation is more likely for the high-elevation species
Predictions
A threshold transformation is more likely for the high-elevation species
Observations
Migration is estimated to be higher for the high-elevation species
Predictions
Migration is estimated to be higher for the high-elevation species
Observations
Maps of genetic diversity correspond with environmental stability in E. iheringii
To sum it up
- Considering the relationship between predicted SDM suitability and local deme size/abundance is important in iDDC models
- High-elevation species are more strongly associated with a threshold transformation compared to low-elevation species and they have higher migration
- This supports the prediction that high-elevation species have wider environmental tolerances than low-elevation species

a Python package to facilitate spatially-explicit coalescent models in msprime
Simulating these complex scenarios is difficult
space
Take spatial models of habitat suitability (i.e. SDMs) projected through time
prime
Simulate the genealogy of a sample of DNA sequences backwards in time with
Baumdicker et al. 2022. Genetics. 10.1093/genetics/iyab229
spaceprime limits coding errors and streamlines analysis
spaceprime: 174,446 events
projections = rasterio.open("data/projections_agg.tif")
locs = gpd.read_file("data/distincta_localities_ex.geojson")
demes = sp.raster_to_demes(projections, transformation="linear", max_local_size=1000)
d = sp.spDemography()
d.stepping_stone_2d(demes, rate=0.001, scale=True, timesteps=1000)
d.add_ancestral_populations(anc_sizes = [100000], merge_time = 23000)
sample_dict, _, _ = sp.coords_to_sample_dict(projections, locs)
ts = msprime.sim_ancestry(demography=d, samples=sample_dict, sequence_length=1e6, recombination_rate=1e-9, random_seed=43, record_provenance=False)
ts = msprime.sim_mutations(ts, rate=1e-8, random_seed=99)msprime: 4 events
demography = msprime.Demography()
demography.add_population(name="A", initial_size=10_000)
demography.add_population(name="B", initial_size=5_000)
demography.add_population(name="C", initial_size=1_000)
demography.add_population_split(time=1000, derived=["A", "B"], ancestral="C")
demography.set_symmetric_migration_rate(populations=["A", "B"], rate=0.1)
ts = msprime.sim_ancestry(samples={"A": 2, "B": 2}, demography=demography, random_seed=12)
ts = msprime.sim_mutations(ts, rate=3e-4, random_seed = 42)With spaceprime you can
- Set up simulations
- Visualize your models
- Analyze your simulation and empirical data
- Run simulations in parallel on a cluster
- Execute these simulations fast
- < 1 hour instead of days
The spaceprime website (will) contain
The spaceprime website (will) contain
- Documentation of all functions
- A quickstart guide and worked example(s)
![]()
- A guide for R users
- FAQ and other resources
In conclusion
spaceprimeis convenientspaceprimeis fastspaceprimeis accessible- Help me create a logo and sticker!
The influence of the environment on genetic diversity varies across scales
1. What is the relationship between genetic diversity and environmental change in insects?
- GDE peaks in the subtropics and is distinct from vertebrate patterns
2. What can genetic diversity convey about the processes underlying species responses to environmental change?
- High-elevation lizard species may have broader environmental tolerances than low-elevation species
3. How can I make integrative modeling of species responses to environmental change more accessible?
- With
spaceprime
